Abstract
Clustered Regularly Interspaced Short Palindromic Repeats, or CRISPR/Cas9, is a typical approach for creating both genomic and epigenetic alterations in DNA molecules. It is a flexible and easy-to-use genome engineering method. The Nobel Prize was given to two women, Jennifer Doudna and Emmanuelle Charpentier, for their ground-breaking study employing CRISPR-Cas9, sometimes known as the "genetic scissors." This approach has application in diverse domains such as pharmaceuticals and agricultural biotechnology. It is also employed in the study of uncommon diseases, genetic problems, and different infections. Due to the system's low precise gene modification effectiveness, the introduction of different mutations, and off-target events, its varied applications require improvement. The original function of CRISPR-Cas9 technology was to provide immunity to prokaryotes against bacteriophages, viral DNA, etc. but today this system is used to cure many life-threatening diseases, genetic disorders, etc. Owing to its quick development, research on this technology's ongoing expansion is crucial. Human embryos have also been modified using CRISPR-Cas9 technology, resulting in offspring who will inherit the genetic changes made during the experiment. Inheritable genetic modifications have been outlawed due to ethical concerns regarding human germ cells. We have discussed the role of CRISPR in prokaryotes, the ethical concerns associated altering this technology, and finally the uses of CRISPR-Cas9 technology in this work. There is also a brief discussion of the possible use of next-generation sequencing and CRISPR/Cas9 in personalized medicine.
Keywords
CRISPR-Cas9, genome editing, ethical issues, NGS, precision medicine, biotechnology, food industry, cancer, and therapeutics
Introduction
Site-specific gene editing is achieved by CRISPR Cas9 genome editing. Atsuo Nakata and his team at Osaka University in Japan first identified CRISPR in E. Coli in 1987. [1,3] There have been reports of the presence of CRISPR, or clustered regularly interspaced short palindromic repeats, in both gram-positive and gram-negative bacteria. This system defends prokaryotes against viral infections, bacteriophage attacks, and other immune system threats. It is a crucial component of the adaptive immunity of prokaryotes.[1]
The Three main stages (adaptation, expression and interference) of the CRISPR Cas immune responses are –
- Adaptation –The Cas protein complex cut a part of the target DNA, the protospacer, after recognizing the specific motif called the protospacer-adjacent motif (PAM).
- Expression and interference – the protospacer DNA can now act as a spacer and is inserted into the CRISPR array. CRISPR RNA (crRNA) is transcribed and recognizes the PAM sequence from invaded viruses or plasmids. The recognized genome is cleaved and gets inactivated. [1]
CRISPR- Cas system is divided into –
- 2 classes (Class 1 and Class 2) [2]
- 6 types (Type I, II, III, IV, V, and VI) [2]
Class 1 generally includes Type I, III, and IV
Class 2 generally includes Type II, V, and VI [3]
The role of Types I, II, and V is to identify and cleave the DNA.
The role of Type VI is to edit RNA, and Type III is to edit DNA and RNA. [3]
The Cas9 nuclease is also called “genetic scissors” and is the only endonuclease to introduce DNA double-strand breaks (DSBs). The Cas9 protein was first isolated from Streptococcus pyogenes called SpCas9. Cas9 protein contains 2 lobes-
- Target recognition lobe
- Nuclease lobe. It consists of 2 domains that are HNH and RuvC. The function of both of these domains is to cleave one of the target DNA strands. [2] [1] [4].
PAM stands for "Protospacer Adjacent Motif," which refers to the brief motifs that are typically present next to protospacers but were not present in the locus's initial spacers. These patterns are crucial for identification. They indicate that the fragment is exogenous and has to be eliminated when they appear at the conclusion of any sequence. The crRNA (CRISPR-associated RNA) is a critical RNA molecule as it directs the proteins of the immune system toward foreign molecules. Another important RNA molecule is tracrRNA (trans-activating RNA) is essential for nuclease activity.[2] When these two RNA molecules—tracrRNA and crRNA—combine, they create a single guide RNA (sgRNA), which is useful for CRISPR-Cas9 system applications. [2] The short DNA fragment known as PAM follows a 20 base pair segment of the sgRNA that matches the target DNA region. [4,2]
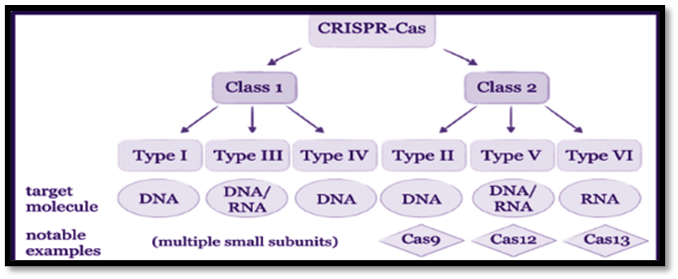
Fig. 1. Conventional classification of known CRISP
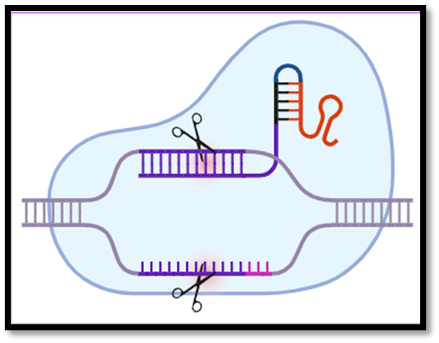
Fig. 2. Structure of CRISPR Cas
Spacers are distinct sequences that are found in between palindromic repeats. These exogenous fragments come from mobile genetic components that have infected prokaryotes, such as plasmids, bacteriophages, etc. In order for the bacteria to remember a previous infection, the virus inserts a tiny fragment of viral DNA into the CRISPR assay when it infects the bacteria.
The Cas proteins used here are Cas1 and Cas 2. CRISPR is a component of the prokaryotic genome and also inspired a method of genome editing in eukaryotes. By providing accurate and effective tools for gene editing, CRISPR-Cas9 has revolutionized the science of genetic engineering. It is a very interesting subject because of its adaptability and possible uses in industries including biotechnology, agriculture, and medicine.
REVIEW OF LITERATURE
Mechanism Of Crispr In Prokaryotes
Any virus, including bacteriophages, can infect bacteria. The virus's DNA enters the cell, joins the bacterial genome, and acts as a spacer between palindromic sequences that are repeated. Every spacer in the system is from a separate virus. Thus, they are referred to as CRISPR arrays because they are positioned between repeating palindromic sequences. Pre-crRNA was created by the transcription procedure applied to this CRISPR array. The Cas9 protein now enters the scene. Here, Cas is an acronym for the protein CRISPR-associated nuclease, which cleaves DNA.
Cas9 along with tracrRNA (they have sections complementary to palindromic sequence) anneals to the palindromic sequences. That means we have a complex that mainly includes pre-crRNA, a tracrRNA, as well as a Cas9 protein. We have another enzyme called RNaseIII that will act on the strands and remove them from in between these complexes thus forming effector complexes (individual crRNA complex). With the help of the effector complexes, the cell can now defend against the foreign elements whose genetic material produces that crRNA.
When the complex comes across a section of viral DNA that matches the sequence of crRNA, the nuclease enzyme identifies and binds to the short unique sequence within the viral genome known as the protospacer adjacent motif (PAM). The function of this enzyme is to cut both strands of DNA a few sequences upstream from the region called PAM. Now the genome of the bacteria can no longer transcribe fully and the infection is impossible. There are numerous biotechnological uses for this phenomena. While crRNA and tracr RNA are distinct molecules in bacteria, they need to be linked together using a linker to generate sgRNA, or single guide RNA, for laboratory purposes. Cas9 and sgRNA combine to produce an effector complex. Thus, in the lab, the proper sgRNA with the complementary sequence is created and added to the cell with the Cas9 protein. This complex reads the DNA and then binds and assembles itself. The complex cleaves DNA and introduces double-strand breaks when it detects the PAM sequence at the 3' end of the target sequence. [1] Natural DNA repair mechanisms will take place following the incision. [1]
DNA repair can occur through two pathways known as NHEJ elongated as nonhomologous end joining and HDR elongated as homology-directed repair. The former pathway is generally observed in eukaryotes. The major function of this NHEJ pathway is to repair double-stranded breaks in DNA. It directly ligates it without the requirement of a homologous template. In this method, no template is required. This pathway generates small changes in the form of insertions or deletions called indels at the cleavage site thus causing frameshift mutations and gene disruptions. [3] It disrupts gene transcription and translation process eventually leading to the knockout of specific genes. [4] This repair mechanism occurs at a very fast rate and is active during the cell
cycle (excluding the process called mitosis). [3] NHEJ produces strands with non- uniformity in sizes. HDR sequence is generally observed in bacteria and archaea. It is a more complex process than NHEJ and uses a homologous DNA template. It mainly uses sister chromatids as a template for its repair.[3] The DNA template which is homologous to the surrounding sequence is utilized to integrate new DNA fragments. This template thus guides the repair mechanism, lowers the chances of errors, and precise knock-in can be achieved.[1] No insertion or deletion is seen in this pathway thus HDR maintains uniformity in the sizes. It generates long sequence mutations.[3] This repair process rarely occurs in nature. [3] HDR process is a slower and more accurate process but is restricted to the S and G2 phases.[3]
To transport the CRISPR and Cas protein complex, both DNA and RNA viruses are involved. The best viruses to deliver are those that belong to plant families, such as the Gemini virus, also abbreviated as GE. GE has a strong effect on a wide range of plants.[3] It results in a greater ratio of CRISPR-Cas mutations in their hosts as well as large quantities of replicons.[3] Haploid-inducer mediated genome editing (IMGE) technology is a recently developed method that typically involves increasing the efficiency and decreasing the breeding time in maize.[3] In an effort to distribute CRISPR-Cas9 efficiently in vivo, researchers are utilizing viral vectors like as adeno-associated virus (AAV). [4,5]
Patent Litigation
The first patent application for the CRISPR technology was filed in May 2012, by both Doudna that was represented by the University of California at Berkeley, and Charpentier, that was represented by the rules of the University of Umeå as an individual inventor.[2] In December 2012 another group Zhang and the Broad Institute applied. [2] In 2019 both parties had patents in this area but in February 2022 the U.S. Patent and Trademark Office Appeal Board confirmed the priority of Zhang and the Broad Institute to be considered the main patent holder for the use of the technology called
CRISPR –Cas9 in human cell line. The dispute in Europe to hold major patents of the technology was won by Doudna and Charpentier in the countries like the U.K., China, Japan, Australia, New Zealand, and Mexico. [2]
Precision Medicine And Next-Generation Sequencing
The goal of precision medicine is creating a personalized treatment plan for each patient. [4] NGS not only helped with individualized care but also with comprehending the ailment and distinct genetic characteristics of the patient. [4] NGS can be used to examine an individual's genetic profile and any alterations in genes that may assist pinpoint potential treatment targets. CRISPR technology can then be used to modify these genes. More effective gene-targeted alterations can be created with the help of the genome editing method called CRISPR.[4]
To determine the treatment decisions and medical approaches the circulating DNA (liquid biopsy), immunological markers, and other biological aspects are analyzed. Various diagnosis methods to determine personalized medicine options are tumor and cell-free DNA profiling, immune markers, RNA analyses, etc. [4]
One of the important reasons for opting for personalized medicine is drug resistance. Many tumors over time develop resistance to chemotherapy due to DNA mutations. NGS (next-generation sequencing) helps in genetic profiling and target identifications. It generally allows the screening of a larger number of genes in a single test. It allows the detection of the mutations that are most frequent to the mutations that occur in fewer than 1% of the patients.[4] The three important steps involved in NGS are- Library preparation and amplification, sequencing, and data analysis. [4] Some of the technologies used to apply NGS to target panels are TAmseq elongated as Tagged- Amplicon deep sequencing, Cancer Personalized Profiling by deep sequencing (CAPP- Seq) as well as Safe-Sequencing System abbreviated as Safe-SeqS.[4]
Among the clinical uses for NGS are the following:
- Biomarkers found through NGS analysis of liquid biopsy samples can be utilized for cancer diagnosis and treatment at any stage of the disease. [4] It is thought to be a more precise method than PCR for locating potential genomic alterations.By identifying unusual mutations in the genome, NGS can assist in distinguishing between them.
- Less than 1% for the minor allele frequency (MAF). NGS is widely utilized to guide decisions about tailored treatment and identify novel biomarkers for early lung cancer detection.
Crispr-Cas: A Case Study In Gene Editing Breakthroughs
In 2015, a group of scientists from Sun Yat-Sen University (Guangzhou, China) published an article based on the usage of the CRISPR-Cas9 system. The researchers stated that they used a fusion of two sperms with one egg which is known as the non-viable embryo and is discarded by in-vitro fertilization (IVF) laboratories.[2] The conclusion was that the technology was not ideal to perform in the embryonic cells of humans.
One of the most important and shocking news about using CRISPR technology was given by a Chinese scientist He Jiankui in November 2018 when he announced the birth of babies that have undergone gene modifications using the CRISPR-Cas9 System. Other scientists were worried that it may create unplanned (‘off-target’) mutations in the genome of the babies that may be a risk. He started his experiment by announcing on an online networking platform named WeChat to recruit volunteers that are married and want to modify the genetics of their children to become resistant to the human immunodeficiency virus (HIV). Some of the requirements he put forward to participate in this experiment were-
- The couple must have a university degree and the man should be HIV positive and the woman to be HIV negative. This was because the risk of transmitting the virus will be minimal.[2]
He planned to modify the cell surface receptor CCR5 gene through binding to which the virus enters the cell. Out of all the responses and further consultations seven couples were selected for the IVF procedure with an additional step of genome editing. After several unsuccessful attempts, two pregnancies led to the birth of babies
The initial pregnancy caused the birth of girls, Lulu and Nana that were twins. The information about the second pregnancy is not known.
It was observed that in Nana both the copies of CCR5 gene were inactivated while in Lulu one of the copy was modified. This shows that only Nana has a chance to be protected from HIV infection. [2]To Lulu, this treatment did not provide any protection because even one of the copies of the gene is enough to produce a receptor on the membrane. These babies were born premature and kept in observation.[2] Some of the scientists that were able to get access to the manuscript of He that were unpublished, found out that many cells were mosaics that were selected for sequencing. This refers to the condition that the information produced during the sequencing of selected cells cannot be considered for the entire embryo as a whole.[2] There were several questionable circumstances of this experiment such as the qualification of He for clinical research, obtaining approval from the ethical committee, possible side effects from loss of a valid copy of the CCR5 gene, and the possibility of introducing off-target mutations thus risking the life of the mother and the babies. This international scandal led to the house arrest of He and then sentenced to prison for 3 years. He is recently released from prison in 2022.[2]
Applications
The first study on human cells using CRISPR technology was done in China in October 2016. This study included the inactivation of the gene PD-1 ex-vivo so that the cells which were modified can attack non-small-cell lung cancer in the patient. The ex-vivo therapy in July 2019 in the U.S. was performed on patients with sickle cell anemia. This therapy improved the condition of patients for at least a few months but the procedure was very expensive (estimated to be around 0.5-1.5 million U.S. dollars) thus acting as an obstacle for widespread use of the technology. [2]
For Crop Improvement
With the growth in the human population, the demand for food production has also grown by almost 70%. The introduction of advanced technologies like CRISPR can help with improvement and increase the production of crops.[3] CRISPR technology helps in editing the specific genes and can be delivered directly as ribonucleoprotein complexes also called RNP complexes containing the Cas protein and the guide RNA.
RNPs also help in reducing off-target mutations or effects in crops. Cis-regulatory regions in DNA sequences are the non-coding regions [20]. These domains consist of CREs which may have variations in their genetic makeup that in turn vary the genetic expressions in crops such as single nucleotide polymorphism, insertions, deletions etc. Phenotypic diversity in crops can be achieved through genetic engineering in these cis- regulatory cites [22]. For instance, an SNP in the promoter region of MYB31 leads to stronger binding of WRKY9 which leads to peppers (Capsicum chinense) being more pungent [21].
Some of the evident benefits of the use of CRISPR technology in major crops are as follows-
Maize (Scientific name is Zea mays)-The technology CRISPR was used to produce hybrids of maize plants that were high in amylopectin. The gene responsible for the regulation of phytic acid synthesis is maizZmIPK. This was also edited by the technology resulting in higher efficiency (13.1%) compared to the samples (9.1%).[3]
Cotton (Gossypiumhirsutum)- The development of lateral roots helps in the growth which thus results in higher fiber yield under drought and low fertility conditions. The rice arginase gene also called the OsARG gene shows a negative impact on the regulation process of root surface areas. Thus CRISPR technology was used to knock out this gene on both the chromosomes that were A- and D-. The result shows that the mutant species formed more lateral roots which could lead to higher biomass. [3]
Rice (Oryza sativa)- Unsaturated fatty acids are essential for the structure and operation of cellular membranes. Fatty acid desaturase 2 (FAD2) is the primary enzyme that catalyzes the transformation of oleic acid into linoleic acid. We can produce rice species with high oleic acid content but low linoleic acid content by deleting the OsFAD2-1 gene. As a result, the quantity of linoleic acid dropped to undetectable levels while the amount of oleic acid increased by 200%. [3,1]
Tomato (Solanumlycopersicum)- a non-proteinogenic amino acid known as gamma-aminobutyric acid (GABA) is present in a very high concentration in tomatoes. The regulatory enzyme in GABA biosynthesis is identified as glutamate decarboxylase.[3] In order to increase the production of GABA, the C-terminal domain that was auto-inhibitory in the enzyme glutamate decarboxylase was edited using CRISPR Cas. It led to the production of high-GABA accumulation tomatoes and significantly improved the blood pressure-lowering function of tomato fruit.[3] It was discovered that editing the gene ALC in tomatoes using the CRISPR Cas system can delay the color change and early ripening without affecting the maturing of fruit and harvest time. [3]
Japanese Luxury Melon (Cucumis melo var. reticulatus "Harukei-3")- Ethylene hormone in plants facilitate ripening of fruits. Enzyme 1-aminocyclopropane- 1-carboxylic acid oxidase (ACO) is involved in the last step prior to ethylene production in melon. Although melon contains five CmACO genes, only CmACO1 is expressed in the fruit at post-harvest stage. Mutation in CmACO1 gene has been done using CRISPR-Cas9, by following the agrobacterium-mediated transformation procedure [14], to inhibit ethylene production in the fruit which showed a potential result on experimentations [13].
For Food Industry
Industrial microbes like Lactobacillus acidophilus, Lactobacillus casei, etc. are ideal targets for the CRISPR-Cas system. [3]These microbes play many important roles like enhancing nutrients and organoleptic qualities of food, preserving food, and so on. [3] Microorganisms in starter cultures generally contain a higher amount of CRISPR-Cas as their adaptive immune system. [3] This system is utilized to enhance traits like survival rate through the gastrointestinal tract, colonization on the host, resistance to bile, etc. [3]
To identify specific genotypes of Streptococcus thermophilusin a heterogeneous population, CRISPR technology is used as a screening tool. This method is used to select unexpected mutations, control specific traits, and improve starter culture phenotype. The well-known microorganism that is used to produce bio-based chemicals is Saccharomyces cerevisiae. It is generally an ideal microorganism because it grows well at 30°C, but during the fermentation process, it produces heat that escalates the production cost. The CRISPR-Cas system was employed to enable the thriving of Saccharomyces cerevisiae(strain T8-292) at 39°C and the genetically modified strain exhibit excellent cell viability at acidic conditions and increased amount of ethanol concentration.
For Biofuel Production
Because they provide security, sustainability, and a decrease in greenhouse gas emissions, biofuels are gradually replacing fossil fuels. The CRISPR system is employed to enhance the production of biofuels. It is challenging to introduce heterologous CRISPR-Cas systems into prokaryotes because they have their own native CRISPR system.In order to address this issue with the CRISPR-Cas system, multiplex genome editing was proposed. The first successful report of it was in yeast. [3] A number of methods for creating CRISPR for multiplex genome editing were presented.[3] One of the crucial microbes utilized to produce biofuel is Escherichia coli. By using CRISPR and CRISPRi technologies to alter E. coli, the compound 1,4-butanediol was made possible. This was done through an artificial pathway that was regulated by six genes named cat1, cat2, bdh,bldas well as sucD and 4hbd. As a result, the engineered E.coli produced 0.9g/L of 1,4-butanediol in 48 h and when using CRISPRi technology the 1.8 g/L (100% increase) in the production of 1,4butanediol was observed by reduction of gamma-butyrolactone and succinate in the 1,4-butanediol pathway. [3] In cellular metabolism, the phosphoenolpyruvate carboxylase (PEPC) gene is crucial. Researchers Kao and Ng [3] edited the PEPC1 gene in Chlamydomonas reinhardtii using the CRISPRi technology. They noticed that despite having little chlorophyll, CrPEPC1 mutants show high levels of biomass and lipid concentration. Algae, also referred to as the third generation of biofuel because of their high lipid content, are a prominent contender for biofuel production. Using the CRISPR method, the ELT gene—which is in charge of fatty acid breakdown in Chlamydomonas reinhardtii—was deleted. The mutants showed a higher accumulation of lipids i.e. 28% of dried biomass.[3] Another green algae Tetraselmis sp. (Platymonas) is a source of biomass and lipid production. [3] The AGP gene elongated as the ADP-glucose pyrophosphorylase was edited using complexes called CRISPR/Cas-RNP which were used to inhibit the carbohydrate synthesis in the pathway of Tetraselmis sp. The results showed that the content of the lipid was increased by 21.1% and 24.1% in the two AGP genome-edited lines.[3] cAMP receptor protein in E.coli has been the target for CRISPR-Cas9 for several research which showed enhancement in biofuel tolerance [24] and Oxidative stress tolerance [25]. In Z.mobilis, NADH dehydrogenase and ubiquinol peroxidase have been the targets which yielded increased thermotolerance and biofuel production [26]. Cel 6B in Thermobifida fusca was targeted to enhance cellobiose resistance [27].
Clinical Applications Of Crispr
The fusion of the technology called CRISPR along with immunotherapy techniques such as Chimeric Antigen Receptor (CAR) T cell therapy holds the potential for success in cancer treatment. [4] Autologous CAR T-cells have drawbacks like time intensive, cost, and challenges like collecting excellent quality and quantity of T-cells from patients. So the CRISPR-modified allogenic T-cells have the potential to overcome these limitations. [4] Eliminating the PD-1, PDL-1 or CTLA-4 genes is seen as a successful approach to overcome T-cell-based adoptive therapy tolerance in cancer treatment. In 2016, PD-1-deleted T cells were engineered and injected into patients suffering from non-small cell lung cancer. The results were encouraging stating that the survival median of 42.6 weeks was satisfactory and off-target events were only 0.05%. The tumor progressed in all the patients by the end of January 2020. Out of 12 patients, 11 of them passed away as a consequence of tumor development. The one that was remaining underwent treatment. The patient did not die due to the ongoing treatment. [4] Some of the potential limitations in the manufacture of CRISPR/Cas9-edited CAR T cells are decreased lymphocyte viability following electroporation and gene alteration that can cause genetic instability. It can thus lead to genetic alterations at unfavorable positions in the DNA [4]. Current cancer treatments include T cell isolation from the patient's body, followed by CRISPR-Cas9 mediated gene editing and in vivo transfer of the modified cell. This method comes with significant limitations that include preservation cellular integrity and viability. Addressing these issues, several studies have been conducted that aimed at in vivo transfer of CRISPR-Cas9 components. One of them employed nanovesicles derived from protoplast of bacteria. These nanovesicles were first modified to target tumor-associated macrophages (TAMs) in patient body. Further, two CRISPR-Cas9 components were loaded into the nanovesicles: A Cas9- sgRNA ribonucleoprotein targeting Pik3cg, and bacterial CpG-rich DNA fragments that are TLR9 ligands. This strategy modifies the tumor-microenvironment, thus resulting in its depletion in the tissue. The size and charge of CRISPR-Cas9 components pose a barrier to condense them into smaller vesicles, hence biological vesicles have proven their efficacy in it [11]. Vesicles derived from mammalian cells have also been put into use but their involvement in undesired physiological processes in patient body have limited their usage [12], thus proving bacteria-derived vesicles to be potential alternative to safe and efficient transfer of CRISPR-Cas9 components. CRISPR-Cas9 system has proven to be potential in identification and extensive of genetic variants causing diseases such as Late-Onset Alzheimer's Disease (LOAD) [15]. A previously discovered variant of SHARPIN, rs572750141 (G186R), was transferred to HEK293 cells which were cultured to produce a cell line that had suppressed NF-KB pathway. This experiment facilitated study of the expressed mutant proteins in the knock-in cells. There was significant upsurge in the level of A?40 and AB42 secretion into the extracellular fluid. Another study employed knock-down of SHARPIN in macrophages derived from THP-1 which showed similar results wherein phagocytosis of A? by modified macrophages was levelled down [16]. Mechanisms that are responsible for resistance to chemotherapy as well as CRISPR-Cas9 has been studied in a recent article [9]. Gene regulatory networks were used with the help of FSSEM tools to identify the genes associated with MDR cancer cell lines, CRISPR resistant cell lines and over-expressed genes in non-resistant cell lines. Profiles of genotype and transcriptome were used for analysis. It was discovered that some genes that had a role to play in CRISPR-Cas9 resistance also shared a function in transcription and proliferation regulation in these cancer cells.
In oncolytic virus production
CRISPR-Cas9 can also be used to produce viruses that lack virulence but are still capable of lysing cancer cells and are known as oncolytic viruses. [4] Example of such a virus is the production of herpes simplex virus type 1 variant that contains lytic capabilities but genes associated with virulence such as ICP34.5 also called neurovirulence gene and ICP6 (UL39) also known as (ribonucleotidereductase) genes are deleted. The most common defect in cancer is the inactivation of the tumor suppressor gene that is p16INK4A gene along with the inactivation of ICP6 which offers replicative selectivity for cells. [4] To limit replication and promote safety in wild-type cells, the E1A gene has been removed from the oncolytic adenovirus.[4]
Oncolytic myxoma viral (MYXV) gene-editing vector system had been produced with the motive to target activated RAS genes contained in Embryonal rhabdomyosarcoma tumors [17]. Although the exact procedure of RAS signaling in ERMS tumor growth is yet unknown but its role in it of established knowledge which formed the basis of this experiment. RAS mutant cancer cells were hence produced with RAS genes knocked- out. The study revealed a significant decrease in mutant cancer cell viability and increase in their apoptosis and a significant increase in the percentage of cells arrested in the G1 phase of cell cycle. Earlier it was speculated that STING had a role in T-VEC resistance in melanoma cell lines. CRISPR-Cas9 technology was used to knock out STING in these sample melanoma cells [18]. Results , as speculated showed the knock- out cells became sensitive to Talimogene laherparepvec (T-VEC) which is an oncolytic herpes simplex virus, type 1 (HSV-1) and has been approved for the treatment of melanoma [19].
Deactivated Cas9 present in CRISPR
Deactivated Cas9 that is present in cancer is catalytically inactive and can be mobilized by gRNAs at precise target sites on DNA. They generally activate and repress specific target genes when linked to transcriptional activators or inhibitory domains.[4] By targeting epigenetic regulatory machinery, cancer treatment outcomes can be improved. [4,2] In a study by Abraham KJ et al [4] To establish the role of RNA polymerase II in ribosomal synthesis, the knockout tool dCas9 was utlised. To activate and suppress the expression of target genes, combining dCas9 with distinct transcription regulatory domains CRISPR activators (CRISPRa) and CRISPR inhibitors (CRISPRi) are generated.[4]
Crispr/Cas9: Potential Drawbacks
CRISPR technology is the leading technology in editing a wide range of genomes.[3] It is a promising tool for the field of agriculture, environment, and also in clinical therapeutics.[3] There are several concerns degrading the clinical utilization of CRISPR that generally involves effectiveness and safety evaluations, the suitability of modified cells, methods involved in delivery, and examination of off-target effects. It is also observed that CRISPR/Cas9 can induce p53 mutation i.e. p53 can spontaneously mutate, and Cas9 can trigger p53 mediated DNA damage response.[4,5] Other major concerns with this technology are the restricted on-target efficiency, partial editing, and the implication of the future of modified organisms. Another ethical issue is modifying human embryos to cure diseases or prevent them by using this technology. Several reports have highlighted the potential risk to whole genome integrity because of mutations that are by-products of off-target changes due to this technology that can lead to chromosomal alterations, several health problems, etc. [3]
Some occurrences of CRISPR-Cas9 side effects, such as larger on-target structural variations (SVs) and off-target mutations, have been reported. These inconsistencies can be, at least in part, explained by variations in experimental variables like the concentration of Cas9, the delivery strategy, or particular characteristics of the cells being studied.
CONCLUSION
CRISPR/Cas systems have offered a versatile, user-friendly molecular framework for accurately altering and regulating the genetic makeup of organisms across many sectors, speeding up genetic research, and the development of gene therapy for the treatment and curing of disease. It includes a discussion on a controversial experiment involving the genetic modification of human embryos aimed at conferring resistance to HIV through the mutation of the CCR5 gene, which ultimately led to the birth of babies with edited genomes and the imprisonment of the scientist responsible, He Jiankui.
CRISPR-Cas technologies have also increased the crop products value and increased the resistivity property of crops against drought, herbicides, and insecticides agriculture sector. [6] This technology has also helped in biofuel production by genome editing and reprogramming the metabolic pathways of various microorganisms. With further progress in science and growth in the studies of CRISPR technology, it will become more controllable, efficient, and precise which can then make valuable contributions to the field of genome editing. CRISPR Cas system and cancer personalized medicine are still developing and in the earlier stages of research.[8] The genes that are identified by Next Generation Sequencing can be altered using the CRISPR-Cas9 system. Next Generation Sequencing can identify the mutations that are not identified by other means. Thus CRISPR-Cas9, both in genome editing and other sectors of science holds great promise as an evolving and cutting - edge technology.[3] the paper examines the transformative potential and ethical considerations of CRISPR technology in both medicine and agriculture, illustrating its profound implications and the challenges it poses for the future of genomic editing.
REFERENCE
- Chen S, Chen D, Liu B, Haisma HJ. Modulating CRISPR/Cas9 genomeediting activity by small molecules. Drug Discov Today. 2022 Apr;27(4):951966. doi: 10.1016/j.drudis.2021.11.018. Epub 2021 Nov 22. PMID: 34823004.
- Gostimskaya, I. CRISPR–Cas9: A History of Its Discovery and EthicalConsiderations of Its Use in Genome Editing. Biochemistry Moscow 87, 777– 788 (2022). https://doi.org/10.1134/S0006297922080090
- Parsaeimehr A, Ebirim RI, Ozbay G. CRISPR-Cas technology a new era in genomic engineering. Biotechnol Rep (Amst). 2022 Apr 12;34:e00731. doi: 10.1016/j.btre.2022.e00731. PMID: 35686011; PMCID: PMC9171425.
- Selvakumar, S.C., Preethi, K.A., Ross, K. et al. CRISPR/Cas9 and next generation sequencing in the personalized treatment of Cancer. Mol Cancer 21, 83 (2022). https://doi.org/10.1186/s12943-022-01565-1
- Wang, SW., Gao, C., Zheng, YM. et al. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer 21, 57 (2022). https://doi.org/10.1186/s12943-022-01518-8
- Höijer, I., Emmanouilidou, A., Östlund, R. et al. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat Commun 13, 627 (2022). https://doi.org/10.1038/s41467- 02228244-5
- Riesenberg, S., Helmbrecht, N., Kanis, P. et al. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat Commun 13, 489 (2022). https://doi.org/10.1038/s41467-022-28137-7
- J.A. Doudna, E. Charpentier, The new frontier of genome engineering with CRISPR-Cas9, Science (80–) 346 (6213) (2014) 1258096.
- Tomasi, F., Pozzi, M. & Lauria, M. Investigating the mechanisms underlying resistance to chemoterapy and to CRISPR-Cas9 in cancer cell lines. Sci Rep 14, 5402 (2024).
- Mingming Zhao, Xiaohui Cheng, Pingwen Shao, Yao Dong, Yongjie Wu, Lin Xiao, Zhiying Cui, Xuedi Sun, Chuancheng Gao, Jiangning Chen, Zhen Huang & Junfeng Zhang, Bacterial protoplast-derived nanovesicles carrying CRISPR- Cas9 tools re-educate tumor-associated macrophages for enhanced cancer immunotherapy, Nature Communications, article number 950 (2024).


 Diksha Belwal*
Diksha Belwal*
 Debangana Mukherjee
Debangana Mukherjee
 Beenu Kumar
Beenu Kumar



 10.5281/zenodo.11672064
10.5281/zenodo.11672064