Abstract
A series of novel 3-(benzylidene amino)-2-phenylquinazoline-4(3H)-ones were synthesized through the reaction of 2-phenyle-3-amino-3 (H) with benzoyl chloride and compounds containing different substituted aromatic aldehydes, yielding 2,3-disubstituted quinazolines. IR and NMR spectroscopy were performed for characterization of the compounds. Molecular docking studies were conducted using a sophisticated kit that integrates AutoDock Vina software with the structural insights from the sitagliptin crystallographic ligand (PDB ID: 1X70). Detailed analysis of binding energies and interactions with key amino acid residues such as TYR666, GLU205, ASN710, and HIS740 within the active site facilitated the identification of potential lead compounds. These findings are significant for the development of therapeutics targeting type 2 diabetes and related metabolic disorders, offering promising avenues for precision medicine. This research underscores the efficacy of in silico methods in identifying and optimizing new heterocyclic compounds for diabetes management. The findings provide a foundation for the synthesis and biological evaluation of these quinazoline derivatives, paving the way for novel antidiabetic drug development and contributing to the broader field of heterocyclic compound research.
Keywords
Diabetes, Quinazole, Molecular Docking, Crystallographic ligand etc
Introduction
DIABETES MELLITIS:
Diabetes mellitus is a spectrum of conditions that includes hyperglycemia as a common way of detection. Diabetes was once thought as a single disease, but it is clearly a heterogeneous group of disorders that are secondary to various genetic predispositions and precipitating factors. Several pathogenic processes are involved in the development of diabetes. These range from autoimmune destruction of the cells of the pancreas with consequent insulin deficiency to abnormalities that result in resistance to insulin action. Deficient insulin action results from inadequate insulin secretion and/or poor tissue responses to insulin at one or more points in the complex pathways of hormone action. Impairment of insulin secretion and defects in insulin action frequently coexist in the same patient and it is often unclear which abnormality, if either alone, is the primary cause of the hyperglycemia.
Types of Diabetes Mellitus:
- Type-1 Diabetes Mellitus (T1DM)
Immune-mediated diabetes type 1 diabetes, or juvenile-onset diabetes, results from a cellular mediated autoimmune destruction of the B-cells of the pancreas. Markers of the immune destruction of the B-cell include islet cell autoantibodies, autoantibodies to insulin, autoantibodies to GAD (GAD65), autoantibodies to the tyrosine Phosphatases 1A-2 and usually more of these autoantibodies are present in 85-90% of individuals when fasting hyperglycemia is initially detected. The rate of ?-cell destruction is quite variable, being rapid in some individuals (mainly infants and children) and slow in others (mainly adults). Some patients, particularly children and adolescents, may present with ketoacidosis as the first manifestation of the disease. Others have modest fasting hyperglycemia that can rapidly change to severe hyperglycemia and/or ketoacidosis in the presence of infection or other stress.[9] Autoimmune destruction of B-cells has multiple genetic predispositions and is also related to environmental factors that are still poorly defined. Although patients are rarely obese when they present with this type of diabetes, the presence of obesity is not incompatible with the diagnosis. These patients are also prone to other autoimmune disorders such as Graves' disease, Hashimoto's thyroiditis, Addison's disease, vitiligo, celiac sprue, autoimmune hepatitis, myasthenia gravis and pernicious anemia.
- Type-2 Diabetes Mellitus:
Type 2 diabetes or adult-onset diabetes, encompasses individuals who have insulin resistance and usually have relative (rather than absolute) insulin deficiency. At least initially and often throughout their lifetime, these individuals do not need insulin treatment to survive. There are probably many different causes of this form of diabetes. Although the specific etiologies are not known, autoimmune destruction of ?-cells does not occur. This type of diabetes accounts 90-95% of world non-insulin dependent diabetes of those with diabetes. It usually arises in association with the stress of another illness such as infection. This form of diabetes frequently goes undiagnosed for many years because the hyperglycemia develops gradually and at earlier stages is often not severe enough for the patient to notice any of the classic symptoms of diabetes. Such patients are at increased risk of developing microvascular and micro vascular complications. Whereas patients with this form of diabetes may have insulin levels that appear normal or elevated, the higher blood glucose levels in these diabetic patients would be expected to result in even higher insulin values had their B-cell function been normal. Thus, insulin secretion is defective in these patients and insufficient to compensate for insulin resistance. The risk of developing this form of diabetes increases with age, obesity and lack of physical activity.
MATERIAL AND METHODS:
MATERIALS:
Mumbai, Merck specialities private limited, Loba chemicals Mumbai and Samar Chemical India.
The solvents and reagents were of AR grade and some were LR grade purified before use. The silica G (60-120 mesh) used for Thin Layer chromatography (TLC) was obtained from Merck India Ltd. Mumbai. The commercially available grade of solvents and reagents were found to be of adequate purity.
METHODS:
Identification and Characterization:
The synthesized compounds were identified and characterized by following methods:
Thin Layer Chromatography (TLC):
TLC is an analysing method which is used to determine the number of components in a mixture, identity of compounds and the purity of a compound. By observing the appearance of a product or the disappearance of a reactant, it can also be used to monitor the progress of a reaction. TLC is a sensitive technique can be analyzed microgram (0.000001 g) quantities and it takes little time for an analysis (about 5-10 minutes). It is based on the Rf value since different compounds have different Rf values. TLC plates of silica gel G were prepared by spreading method. These were dried in air and then activated by heating in hot air oven at 110?C for 30 minutes. The solvent toluene: ethyl acetate: formic acid (5:4:1, v/v/v) and benzene: acetone (9:1, v/v) used as solvent systems. The spots were located by exposure to iodine vapours or under UV-light.
Melting Point Determination
The melting point of organic compound was determined by open capillary tube method. The determination of melting is the most important and easy way of differentiating the physical constant of one compound from another.
Infra-Red Spectroscopy (IR):
An important tool of the organic chemist is Infrared Spectroscopy. IR spectra are acquired on a special instrument, called an IR spectrometer. IR is used both to gather information about the structure of a compound and as an analytical tool to assess the purity of a compound. IR spectra are quick and easy to run. Infrared radiation is absorbed by organic molecules and converted into energy of molecular vibration. In IR spectroscopy, an organic molecule is exposed to infrared radiation. When the radiant energy matches the energy of a specific molecular vibration, absorption occurs. Thus, IR spectra of each and every bond will be formed. The infrared spectra compounds were recorded on IR Affinity-1 SHIMADZU Ltd. (range 4000500 cm-1) spectrometer.
Nuclear Magnetic Resonance Spectroscopy (NMR):
Nuclear Magnetic Resonance (NMR) spectroscopy is an analytical chemistry technique used in quality control and research for determining the content and purity of a sample as well as its molecular structure. Nuclear magnetic resonance (NMR) is a physical phenomenon in which nuclei in a magnetic field absorb and re-emit electromagnetic radiation. The principle behind NMR is that many nuclei have spin and all nuclei are electrically charged. If an external magnetic field is applied, an energy transfer is possible between the base energy to a higher energy level (generally a single energy gap). The energy transfer takes place at a wavelength that corresponds to radio frequencies and when the spin returns to its base level, energy is emitted at the same frequency. The signal that matches this transfer is measured in many ways and processed in order to yield an NMR spectrum for the nucleus concerned.
SYNTHESIS OF COMPOUNDS:
STEP -1: Synthesis of 2-phenyl-4H-benzoxazin-4-ones:
Anthranilic acid or 6-methyl anthranilic acid (0.1 mol) dissolved in pyridine (30 mL), a solution of benzoyl chloride (0.02 mol) was added in this solution in dropwise manner with constant stirring at 2-8°Cover the period of 1 h. The reaction mixture was further stirred for half an hour at room temperature and treated with aqueous sodium bi carbonatee to remove the unreacted acid. A solid product obtained, which was filtered, washed with cold water to remove the inorganic materials and then with n-hexane and recrystallized from ethanol.
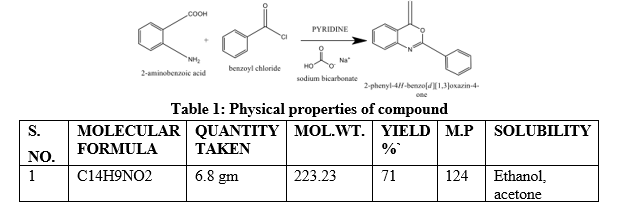
Table 1: Physical properties of compound
STEP-2: Synthesis of 3-amino-2-phenyl-quinazolin-4(3H)-one.
2-phenyl-4H-3, 1-Benzoxazin-4 one (1, 2) was converted to 3-amino-2-phenyl-quinazolin4(3H)-one under reflux for 6-7 hr with hydrazine monohydrate in ethanol. The concentrated solution was poured into ice cold water to obtain a solid product. The obtained solid was washed with water, filtered, dried and recrystallized from ethanol.
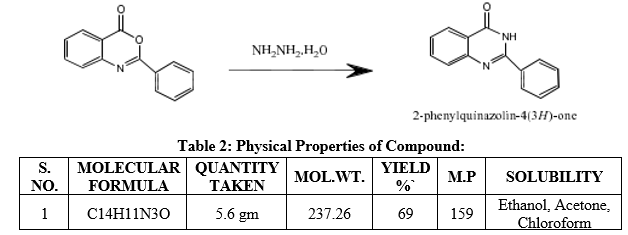
Table 2: Physical Properties of Compound:
STEP-3: Synthesis of Schiff base-
A mixture of 3-amino-2-phenyl-3H-quinazoline-4-one and the appropriate aromatic aldehydes in ethanol (60 mL) containing few drops of glacial acetic acid was heated under reflux for 10 hr. The solution was cooled and the precipitated solid obtain was filtered, washed with water, dried in vacuum and recrystallized from ethanol.

Compound- 2-a

Compound code:
2-a
Molecular formula:
C21H14N403
Molecular weight :
370.36
Solubility:
Ethanol, Chloroform, Acetone
Chemical Name:
3-[(3-Nitro-benzylidine)-amino]-2-phenyl-3H-quinazolin-4 one
Rf value:
0.66
Characterization
Table 3: IR (cm-1,KBr)-

- C
???????

Compound code:
2-b
Molecular formula:
C21H14ClN30
Molecular weight:
359.81
Chemical Name:
3-[(4-Chloro-benzylidine)-amino]-2-phenyl-3H-quinazolin-4 one
Rf value:
0.51
Characterization:
Table 4: IR (cm-1, KBr)

Compound: 2C

Compound code:
2-C
Molecular formula:
C21H15N3O
Molecular weight:
325.36
Chemical Name:
3-(Benzylidine-amino)-2-phenyl-3H-quinazolin-4 one
Rf value:
0.56
Characterization:
Table 5: IR (cm-1, KBr)-

1H NMR (CDCl3, 500 MHz) ? (ppm): 9.04(s,1H,N-C=N),8.071(s,H-5),7.74(H-6),7.70(H-
7),7.75(H-8),8.13-8.15(d,H-3”/H-5”),8.05-8.07(d, H-2”/H-6”),7.68(H-6’),7.63(H-2’),7.57(H-
5’/H-4’).
Compound code
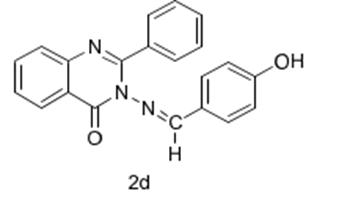
2-d
Molecular formula:
C21H15N3O2
Molecular weight:
341.36
Chemical Name:
3-[(4-Hydroxy-benzylidine)-amino]-2-phenyl-3H-quinazolin-4 one
Rf value:
0.48
Characterization:
Table 6 :IR (cm-1, KBr)-

5. Compound: 2-e
Compound code:

2-e
Molecular formula:
C21H14N403
Molecular weight:
368.43
Chemical Name:
3-[(4-dimethyl benzylidine) amino]-2-phenyl quinazolin-4(3H)-one
Rf value:
0.63
Characterization:
Table 7:IR (cm-1, KBr)-

MOLECULAR DOCKING:
PROCEDURE:
Molecular docking of prepared compounds was performed on one of the enzymes, DPP-4, which represents an effective target in antidiabetic activity. Inhibition of this enzyme results in reduced blood glucose levels. The software used for molecular docking of synthesized compounds was PyRx which uses Autodock Vina for docking based virtual screening. The molecular visualization programs used were PyMol and Autodock Tools.
1. Retrieval of Protein Structure
3D structure of the above enzyme was retrieved from RCSB-Protein Data Bank. The protein data bank was searched for “Dipeptidyl peptidase 4” and among the results, PDB ID: 1X70 (2.1Å) was chosen on the basis of its structure validation criteria.
2. Finding the Active Site of the Protein
The amino acid residues around 4Å radius of bound ligand were chosen as the active site. Some of the important amino acids responsible for DPP-IV function are SER630, HIS740 and ASP708 (the catalytic triad), GLU205, GLU206, TYR547, TYR662 etc. Among all these interactions, the bound ligand (Sitagliptin) exhibited only H-bond interactions in the active site with amino acids: GLU205, GLU206 and TYR662.
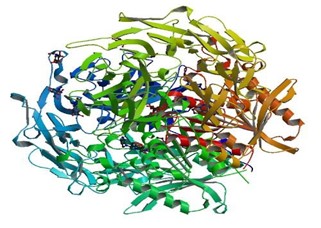
Figure 1: PDB structure of Dipeptidyl peptidase- 4
5. Separation of Protein and Ligand
Next step was to remove the ligand from active site of protein. For this purpose, molecular visualization program PyMol was used. The PDB structure of protein was opened in PyMol and the residue sequence was displayed in the workspace. From here all the hetero atoms were chosen and deleted. This resulted in protein structure without bound ligands. The file was saved.
6. Preparation of Protein
The protein structures retrieved from PDB contain crystallographic waters and do not contain hydrogen atoms. Therefore, for effective docking water molecules have to be removed and polar hydrogens have to be added to the protein structures. This was achieved with Auto dock Tools 1.5.6. Auto dock Vina also requires dimensions of grid as it uses the grid docking methodology. The grid dimensions were set such that they covered entire active site. These dimensions were noted and the structure was saved in pdbqt format (the file format required by Auto dock for docking).
Preparation of Experimental Molecule Structures
Structures of all the four experimental molecules were drawn in Chemdraw Ultra 10.0. These structures were then directly imported to PyRx (through Open Babel module. Open Babel was used for energy minimization of all the Molecules Using Universal Force Field (UFF). This was followed by conversion of all the molecules to pdbqt format.
7. Docking of Experimental Molecules
The pdbqt files of receptor and all the ligands were used for docking in PyRx through Vina wizard, which directs the user through stepwise procedure. In the Vina wizard receptor and ligands were added clicking “Add Macromolecule” and “Add Ligand” functionalities respectively in menu option “Select Molecules”. This was followed by clicking “Run Vina”. In this step the grid was set according to the active site dimensions and docking operation was started by clicking “Run Vina” option at the bottom of the window.
RESULTS:
- PHYSICAL PROPERTIES OF SYNTHETIC COMPOUND
Table 8: Physical Properties of Synthetic Compound

- SOLUBILITY PROFILE OF SYNTHETIC COMPOUND:
Table 9: Solubility Profile of Synthetic Compound

(++)=very soluble, (+) =soluble, (+-) =sparingly soluble, (-) =Insoluble
- ANALYSIS OF MOLECULAR DOCKING:
After the completion of docking run, results were observed using “Analyze Results” pane in Vina wizard. The corresponding table was saved in .csv format to be further analyzed in Microsoft Excel. PyRx saved different output files for each docked ligand as .pdbqt file containing 9 Conformers of same ligand in a single file. The docking protocol was validated by reproducing the binding interactions of crystallographic ligand.
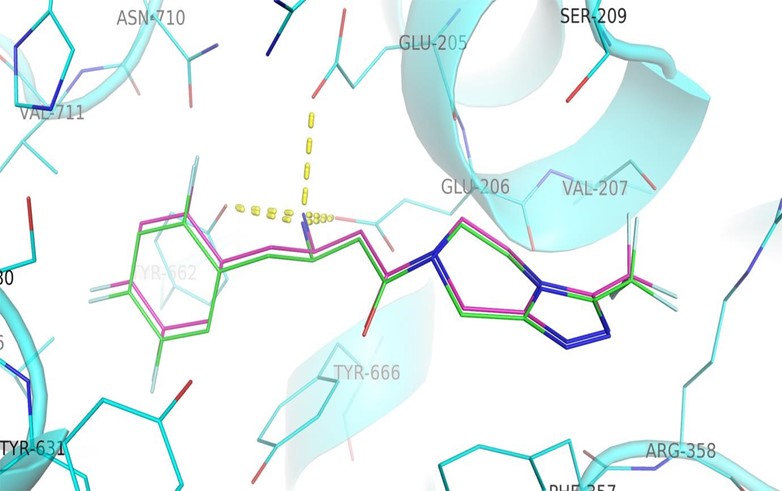
Figure2: Validation of docking protocol: The interactions of crystallographic ligand (violet carbons) were mimicked by docking of ligand structure (green carbons) in Autodock Vina
- INTERPRETATION OF DOCKING RESULTS:
All molecules were docked in the active site of the enzyme DPP-IV. Since Autodock Vina uses a stochastic search algorithm, several docking runs were performed and the best result among all runs is reported in Table 10. This table displays various conformers (modes) of different title compounds (2-a, 2-b, 2-c, 2-d and 2-e) along with their binding affinities and their interactions with target active site. Where the interactions with amino acids represent the conformational search part of the docking process and binding energy represents the scoring part of it.
Table 10: Indicates the molecular docking parameters for compounds 2a-2-e. It demonstrates that compounds have favourable free energy of binding to the receptor.


Figure-11: Compound-02 (A) Interactions with amino acid residues in the active site
(B) Alignment with the crystallographic ligand (Sitagliptin) in the active site

Figure 12: Compound-2a

Figure 13: Compound -2b.

Figure 14: Compound-2c

Figure 15: Compound -2d

Figure 16: Compound-2e
DISCUSSION:
Anthranilic acid reaction with benzoyl chloride yielded 2-phenyl-4H-3, 1-Benzoxazin-4 one by N-acylation via dehydrative cyclization mechanism. Subsequently which was converted to 2-phenyl-3-amino-3(H) quinazoline-4 one with hydrazine hydrate. A series of novel 3 (benzylidene amino)-2-phenylquinazoline-4 (3H)-ones were synthesized by reaction with 2phenyl-3-amino-3 (H) quinazoline-4 one and compounds containing different substituted aromatic aldehyde to afford 2,3 disubstituted quinazoline-4(3H)-one derivatives. The crude compound were crystalized by using ethanol. Purity of the compound were checked by thin layer chromatography. Melting points were taken on a liquid paraffin bath in open capillary tubes.IR and NMR studies were performed for characterization of the compounds. IR (cm-1, KBr)C=O found between (1600-1430),Ar-C-H bending(684.76),C-H Aliphatic (29762850),NO2(1527.16),CCl(1091.57),monosubstituted benzene (900-690) ,H-C=N(1645-1605) and NMR spectra the common peaks 9.04(s,1H,N-C=N), 8.071(s,H-5),7.74(H-6),7.70(H7),7.75(H-8),8.13-8.15(d,H-3”/H-5”),8.05-8.07(d, H-2”/H-6”),7.68(H-6’),7.63(H-2’),7.57(H5’/H-4’).
CONCLUSION:
The synthesis of novel quinazoline derivatives involved a series of reactions beginning with the N-acylation of anthranilic acid with benzoyl chloride, yielding 2-phenyl-4H-3,1benzoxazin-4-one via a dehydrative cyclization mechanism. This intermediate was subsequently converted to 2-phenyl-3-amino-3(H)-quinazoline-4-one using hydrazine hydrate. A series of novel 3-(benzylidene amino)-2-phenylquinazoline-4(3H)-ones were synthesized through the reaction of 2-phenyl-3-amino-3(H)-quinazoline-4-one with various substituted aromatic aldehydes, resulting in 2,3-disubstituted quinazoline-4(3H)-one derivatives. The crude compounds were crystallized using ethanol and their purity was verified by thin layer chromatography. Melting points were determined using a liquid paraffin bath in open capillary tubes. Characterization of the compounds was performed using IR and NMR spectroscopy. The IR spectra showed characteristic peaks for C=O (1600-1430 cm-1), Ar-C-H bending (684.76 cm-1), C-H aliphatic (2976-2850 cm-1), NO2 (1527.16 cm-1), C-Cl (1091.57 cm-1), monosubstituted benzene (900-690 cm-1), and H-C=N (1645-1605 cm-1). The NMR spectra revealed common peaks at 9.04 (s, 1H, N-C=N), 8.071 (s, H-5), 7.74 (H-6), 7.70 (H-
7), 7.75 (H-8), 8.13-8.15 (d, H-3”/H-5”), 8.05-8.07 (d, H-2”/H-6”), 7.68 (H-6’), 7.63 (H-2’), and 7.57 (H-5’/H-4’). The molecular docking studies were conducted using a sophisticated kit that integrates AutoDock Vina software with the structural insights from the sitagliptin crystallographic ligand (PDB ID: 1X70). This approach allowed for efficient screening and evaluation of the synthesized compounds for their DPP-4 inhibitory activity. Detailed analysis of binding energies and interactions with key amino acid residues such as TYR666, GLU205, ASN710, and HIS740 within the active site facilitated the identification of potential lead compounds. These findings are significant for the development of therapeutics targeting type 2 diabetes and related metabolic disorders, offering promising avenues for precision medicine.
ACKNOWLEDGMENT:
The authors convey sincere gratitude to all author for sharing their valuable knowledge and time to create this informative review article.
REFERENCES
- Faisal M, Saeed A. Chemical insights into the synthetic chemistry of quinazolines: Recent advances. Frontiers in chemistry. 2021 Jan 14;8:594717.
- Eweas A, Abdallah Q, Elbadawy M. Synthesis and biological evaluation of some new 2-pyridylquinazoline derivatives. Current Chemistry Letters. 2021;10(4):459-70.
- Tiwary BK, Pathak RK, Pradhan K, Nanda AK, Bothra AK, Chakraborty R. Evaluation of drug candidature of some quinazoline-4–3H-ones as inhibitor of human dihydrofolate reductase enzyme: Molecular docking and In silico studies. Int J Pharm Pharm Sci. 2014;6(2):393-400.
- Jay SS. Synthesis and Biological Evaluation of 1-Aryl-4, 5-dihydro-1H-pyrazolo [3, 4- d] pyrimidin-4-one Inhibitors of Cyclin-Dependent Kinase. Med. Chem. 2004; 47:4113–4117.
- Stuart AR, Eric AG, Minghan W. Chemistry and Biochemistry of Type 2 Diabetes. Chem. Rev. 2004; 104:1255–1282.
- van Belle TL, et al. Insights into beta cell preservation in type 1 diabetes. 2011.
- Brunton SA, et al. Islet function in type 2 diabetes: A review. 2006.
- Craig ME, Stitzel ML. ?-cell death and dysfunction during progression of diabetes. 2004.
- Unger RH. Role of dysfunctional adipose tissue in insulin resistance. 2008.
- Kahn SE, et al. Pathophysiology and treatment of type 2 diabetes: Perspectives on the past, present, and future. 2006.
- Feinglos MN, Bethel MA. Type 2 diabetes mellitus: An evidence-based approach to practical management. 2008.
- Cerasi E, et al. ?-cell dysfunction in type 2 diabetes: A therapeutic target. 2001.
- Haffner SM. Survival as 90% of beta cells are damaged while in T2DM over half the patients lose their response to antihyperglycemics and eventually require insulin therapy. 1998.
- Kahn SE. Main actions of insulin including glycogenesis, enhanced synthesis of fatty acid, esterification of fatty acid, deprived proteolysis, lipolysis, and gluconeogenesis. 1985.
- Sallé A, et al. Drawbacks associated with injectables in diabetes: hypoglycemia, weight gain, and complications. 2004.
- Krentz AJ, Bailey CJ. Glitazones (Thiazolidinediones) mechanism of action and common side effects. 2005.
- Rang HP, et al. Common side effects of Glitazones: weight gain, edema, and anemia. 2003.
- Jiang Y, Zhang Y. T2DM as a progressive disorder, often accompanied by the loss of functions of beta cells, creating the necessity of insulin therapy. 2003.
- Holst JJ, Orskov C. Concept of incretin: the role of gastrointestinal hormones in enhanced insulin release and reduced gastric emptying. 2004.
- Holst JJ, Deacon CF. Physiological regulators for the secretion of GLP-1: response to glucose, fat, acetylcholine, and gastrin-releasing peptide. 2005.
- McKennon S, Campbell RK. Role of dipeptidyl peptidase-4 (DPP-4) in inactivating incretin hormones, GLP-1 and GIP. 2007.
- Baggio LL, Drucker DJ. Role of DPP-4 on GLP-1 inactivation. 2007.
- Verspohl EJ. Effects of GLP-1 on various tissues. 2009.
- Ruozi B, et al. Effects of GLP-1 on various tissues. 2017.
- Hansotia T, Drucker DJ. Glucose-dependent insulinotropic polypeptide (GIP) as an incretin hormone. 2005.
- Wu T, et al. Short half-life of GLP-1 and its inactivation by plasma DPP-4 enzyme. 2009.
- Davidson MH, et al. GLP-1 injectable form associated with side effects and degradation upon oral administration. 2005.
- Croom KF, McCormack PL. Liraglutide, a long-acting analogue of GLP-1, decreases appetite, has no effect on glucagon counter-regulation, and increases beta cell mass. 2009.
- Nauck MA, et al. Incretin-based therapies show benefits such as reduced weight gain, lesser chances of hypoglycemia, and positive effects on beta cell proliferation and differentiation. 2004.
- Fadini GP, Avogaro A. Incretin mimetics and DPP-4 inhibitors have fewer side effects and complications, and show beneficial effects in patients with cardiovascular complications. 2011.
- Drucker DJ. GLP-1 performs various functions like stimulating insulin biosynthesis, inhibiting glucagon secretion, slowing gastric emptying, and reducing appetite, but it is rapidly inactivated by DPP-4. 1998.
- Deacon CF, et al. GLP-1 and GIP get inactivated by DPP4 enzyme, leading to a short plasma half-life. 1999.
- Nauck MA, et al. DPP-4 cleavage site for activated GLP-1. 1996.
- Hopsu-Havu VK, Glenner GG. DPP-4 or CD26 is a serine protease enzyme belonging to the prolyl endopeptidase family. 1966.
- Cunningham DF, O’Connor B. DPP-4 enzyme cleaves dipeptide from the penultimate position from target polypeptides like chemokines and peptide hormones. 1997.
- Iwaki-Egawa S, et al. DPP-4 expression in various cell types including epithelial cells of the intestine, liver, corpus luteum, prostate, and kidney. 1998.
- Deacon CF, Holst JJ. DPP-4 inhibitors and therapeutic evolution in glucose homeostasis. 2002.
- Lambeir AM, et al. Structure of DPP-4 enzyme with catalytic domain and eight-bladed ?-propeller domain. 2003.
- Gorrell MD. Dipeptidyl peptidase family preferentially cleaves dipeptides from the penultimate position having proline or alanine. 2005.
- Abbott CA, et al. The only difference between the DPP family and the prolyl oligopeptidase superfamily is the presence of two glutamic acid moieties within the catalytic site defining the enzymatic potency. 1999.
- Wang B, et al. FAP (seprase) shares 51% similarity in amino acid identity to DPP-4, with similar substrate specificity but different expression patterns. 2008.
- Ajami K, et al. DPP-8 and DPP-9 have 26% similarity with DPP-4, are localized to the cytoplasm, and hydrolyze substrates with less efficiency compared to DPP-4. 2004.
- Rummey C, Metz D. Advantageous active sites of DPP-4 differ from those of DPP-8 and DPP-9, allowing selective inhibition of DPP-4. 2007.
- Thoma R, et al. Prolyl endopeptidases or prolyl oligopeptidase (PEP/PREP or POP) hydrolyze the bond following proline, structurally homologous to the ?/? hydrolase domain of DPP-4. 2003.
- Oefner C, et al. The human counterpart of DPP-4 is made up of 766 amino acids, forming a homodimer with two domains - ?/?-hydrolase and ?-propeller. 2003.
- Jiang J, Ghosh M. Structure of DPP-4 with ?/?-hydrolase domain, ?-propeller domain, active site, and substrate binding site. 2017.
- Tuan-Anh NP, Zunhua Y, Yuanying F, Jun L, Jongkook L, Haeil .Synthesis and biological evaluation of novel 2,4-disubstituted quinazoline analogues as GPR119 agonists. Bioorg & Med Chem. 2013; 21(5): 1349–1356.


 Brijmohan Sahu* 1
Brijmohan Sahu* 1
 Tapas Kumar Panigrahi 2
Tapas Kumar Panigrahi 2




















 10.5281/zenodo.13294557
10.5281/zenodo.13294557